Inferring fitness landscapes and selection on phenotypic states
Inferring fitness landscapes and selection on phenotypic states
Recent advances in single-cell time-lapse microscopy have revealed non-genetic heterogeneity and temporal fluctuations of cellular phenotypes. While different phenotypic traits such as abundance of growth-related proteins in single cells may have differential effects on the reproductive success of cells, rigorous experimental quantification of this process has remained elusive due to the complexity of single cell physiology within the context of a proliferating population.
We have developed a practical empirical method to quantify the fitness landscapes and selection strength on arbitrary phenotypic traits, using genealogical data in the form of population lineage trees. The method provides a unified and practical framework for quantitative measurements of fitness landscapes and selection strength for any statistical quantities definable on lineages, and thus elucidates the adaptive significance of phenotypic states in time series data.
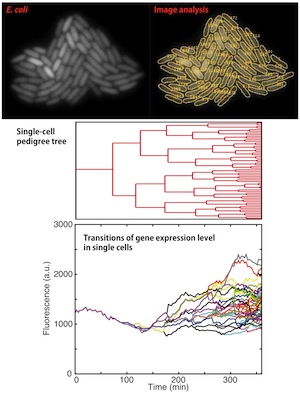
An example of single-cell lineage tree and time-course dynamics of single-cell phenotypes on the lineages.
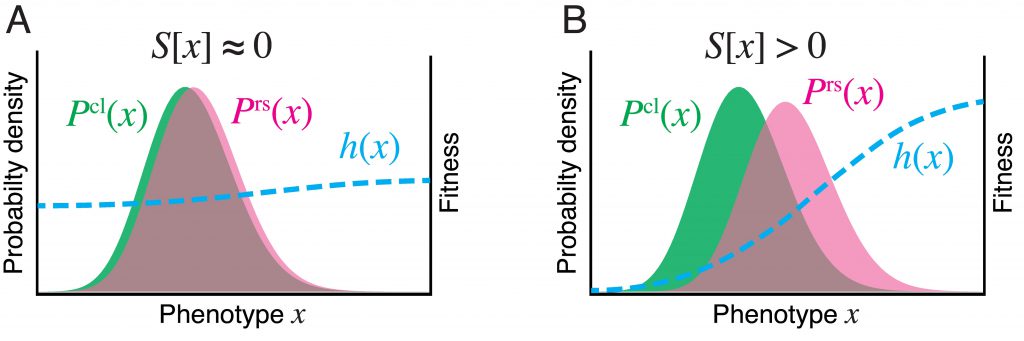
Fitness landscapes and selection strength(From: Nozoe, et al., PLoS Genetics, 2017)
See also:
- Nozoe T, Kussell E, Wakamoto Y (2017) Inferring fitness landscapes and selection on phenotypic states from single-cell genealogical data. PLoS Genet 13(3):e1006653.
- Yamauchi, S., Nozoe, T., Okura, R., Kussell, E., Wakamoto, Y. (2022) A unified framework for measuring selection on cellular lineages and traits. eLife. 11, e72299.

 日本語
日本語